The central goal of my research is to understand how cells work from the micron to angstrom scale. The common thread to my research projects is methods development in structural biology. I have extensive experience in cryogenic electron microscopy (CryoEM), from demonstrated success in solving protein structures by single particle cryoEM, to imaging cells with cryogenic electron tomography (cryoET) and solving protein, peptide and small molecule structures using Microcrystal Electron Diffraction (MicroED). I have also used x-ray crystallography.
I've worked with a wide variety of samples, from ribosomes to membrane proteins. I'm passionate about developing strategies to image small proteins by electron cryomicroscopy using single particle analysis, and to streamline Microcrystal Electron Diffraction (MicroED) experiments. I'm interested in optimizing grid preparation and thinking about how to effectively teach others to do electron microscopy.
Microcrystal Electron Diffraction (MicroED)
Microcrystal Electron Diffraction (MicroED) is a cryoEM technique that can solve atomic resolution structures from small 3D crystals. Crystallography is a difficult art, and many crystals never grow large enough for x-ray crystallography experiments. MicroED can be used to examine these tiny crystals and provide structures. With my deep expertise in cryoEM methods, I joined Tamir Gonen’s lab to develop methods to streamline all steps in a MicroED experiment, from sample preparation, data collection, and data processing.
I thrive in collaborative environments and take great care in developing protocols and training materials to share with in my laboratory and the world. I was invited to speak about MicroED sample preparation for an SB Grid Webinar and you can watch my talk below.
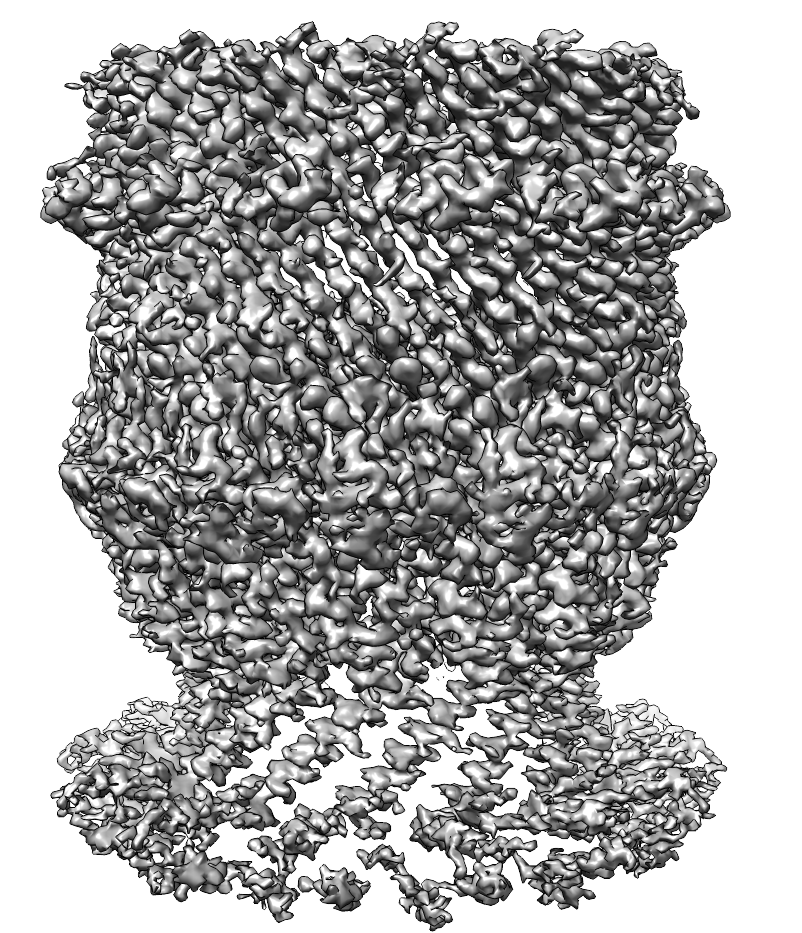
Structure of the Vibrio cholerae Type IV competence pilus secretin PilQ
Horizontal gene transfer facilitates the spread of antibiotic resistance genes among bacterial populations. Natural transformation is a mechanism of horizontal gene transfer. In Vibrio cholerae, the Type IV competence pilus mediates natural competence. In collaboration with Prof. Ankur Dalia (Indiana University), Dr. Triana Dalia (Indiana University), and Prof. Matthew Sazinsky (Pomona College), I purified the V. cholerae secretin PilQ directly from V. cholerae and used single particle cryoEM to solve the structure. Based on the structure, I designed cysteine pair mutants to reversibly inhibit transformation. Learn more about what we discovered by checking out our preprint at bioRxiv and the final paper at Nature Communications!
Single particle cryoEM for small proteins
Small proteins are difficult to identify in micrographs and reconstruct by single particle cryoEM. We developed a large, symmetric cryoEM platform (DARPin-aldolase) to display a small protein and facilitate structure determination. Recently, we posted a pre-print of our paper “Fusion of DARPin to aldolase enables visualization of a small protein by cryoEM” to the bioRxiv. The final version of the paper is published in Structure
Tomography of bacterial secretion systems
How do bacteria acquire antibiotic resistance? Many bacteria release DNA into their environment to share with their neighbors. They also take up DNA from the environment. How does this DNA transfer work, and how can we develop tools to combat it in the case of pathogenic bacteria? Electron cryotomography can help us answer these questions.
This slice from a tomogram of a Vibrio cholerae cell demonstrates the presence of a chemoreceptor array (green) and a flagella motor (pink). The scale bar is 25 nm